|
||
|
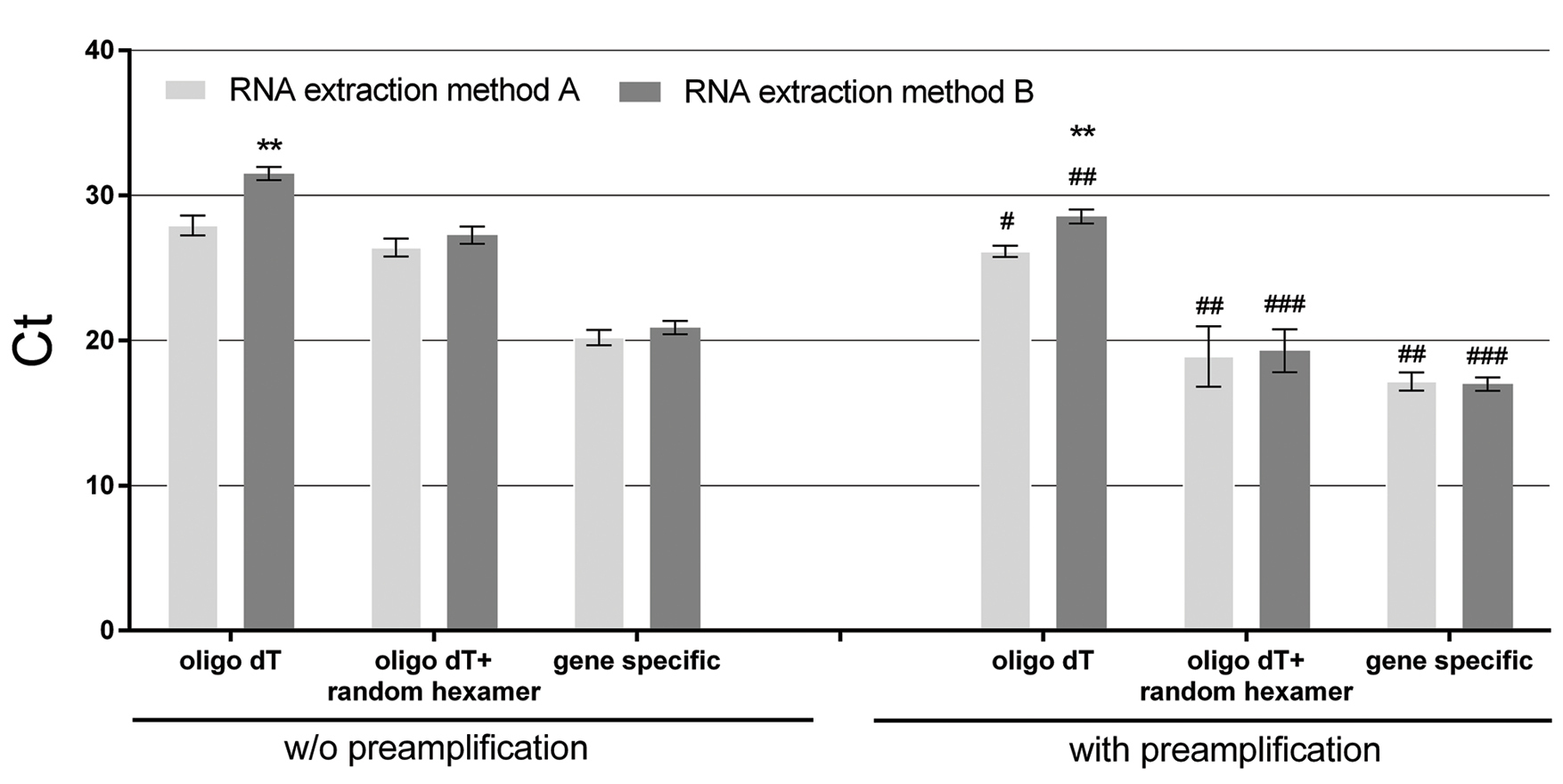
Comparison of Ct values obtained through two-step qPCR of non-preamplified and preamplified cDNA samples. Data are presented as mean ±SD of FFPE tissue samples for both RNA extraction methods utilizing three different reverse transcription priming strategies (oligo dT; oligo dT+random hexamer; gene specific reverse primer). **p<0.01 vs. RNA extraction method A; #p<0.05, ##p<0.01, ###p<0.001 vs. respective priming strategy and RNA extraction method w/o preamplification. A. Lysis buffer RNA extraction (grey); B. lysis buffer and phenol-based RNA extraction (dark grey). |